Access to the National Library of Estonia newspaper and periodical collections
Description
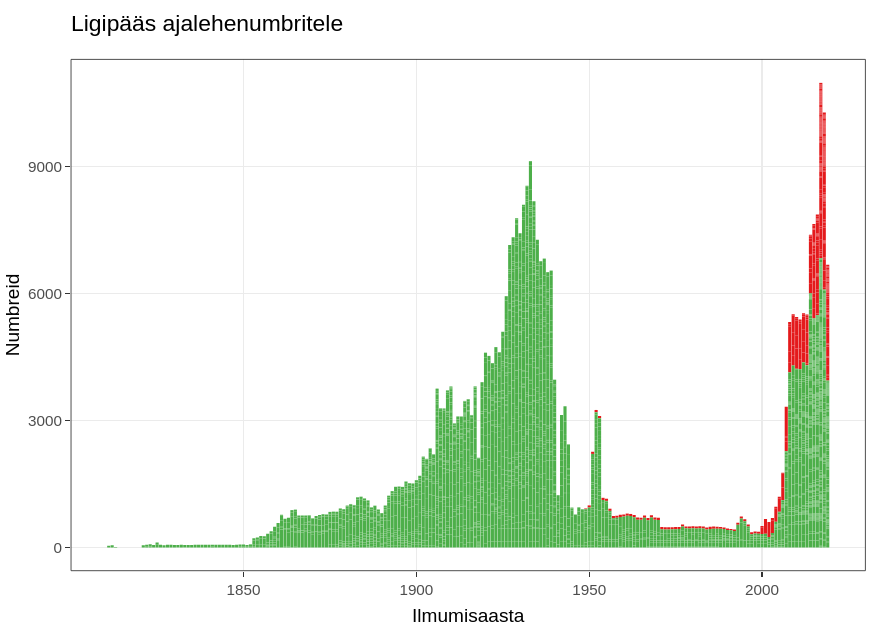
Digitized Articles of Estonia is searchable through the web interface https://dea.digar.ee/ and accessible as a dataset. The overview of the data is on a separate page.
The access to the data is available in the cloud in JupyterHub environment where it is possible to run code and write Jupyter Notebooks, using R or Python.
The JupyterHub environment has access to full texts and metadata, a possibility to write your own analyses and download your findings. Data is open for anyone to use.
How to begin
To use the environment, you need to acquire a username in ETAIS. To get the username please contact data@digar.ee.
For easy access to the data, an R package digar.txts has been made that helps one form subsets from the collection and perform full text searches on them.
To process the data it is possible to use your own code, rely on example case studies or export your search results as a table.
In short
- Go to the webpage https://jupyter.hpc.ut.ee/ and log in (username from data@digar.ee).
- Use the default parameters (1 CPU core, 8 GB memory, 6h timelimit)
- Create a new notebook with an R kernel.
- Use the code from the Using code section
- Upload the example for beginners: english
- Have a look at example case studies: hobu, elekter, aur 20. saj algul (.html, .ipynb, .Rmd)
- Workshops: Nelijärve 5. nov 2020, Kevad 2020 eestikeelne lühikursus tekstitöötlusest R-is
Note
- In the University of Tartu network, it is possible to also access the data via RStudio https://rstudio.hpc.ut.ee/. In the future this chance should become available for all users.
Starting up JupyterHub
- Go to the webpage https://jupyter.hpc.ut.ee/ and log in.
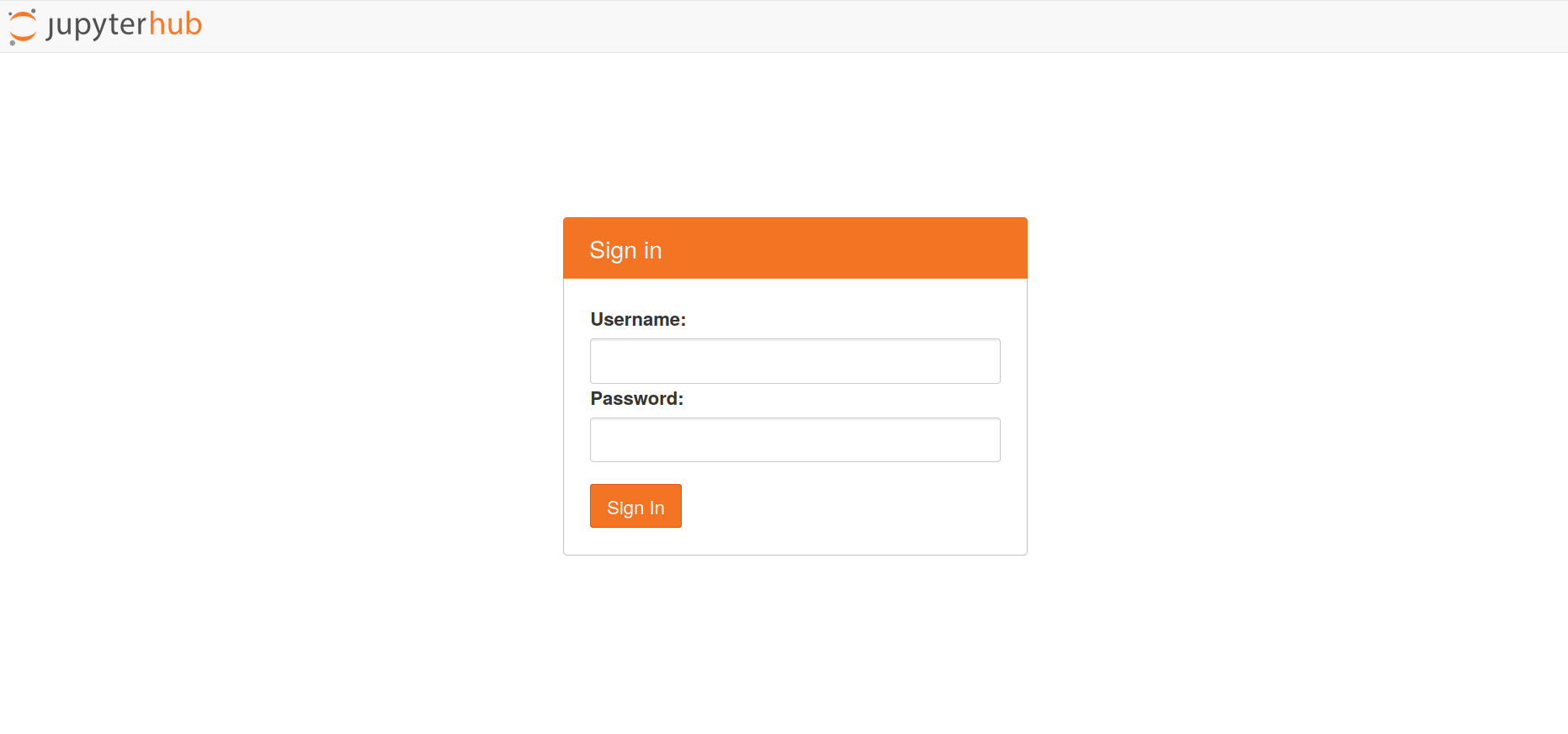
- Pick the fisrt offered option (1 CPU core, 8GB memory, 6h timelimit). This opens up the data processing for 6 hours. All your files will be kept permanently with your username.
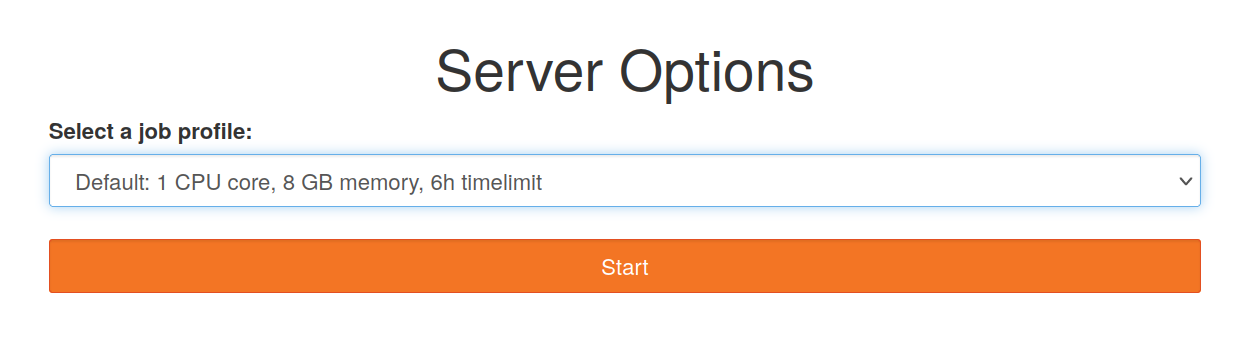
- Wait until the server starts up. This may take a few minutes depending on the current queue. Sometimes refreshing the page in the browser may help.
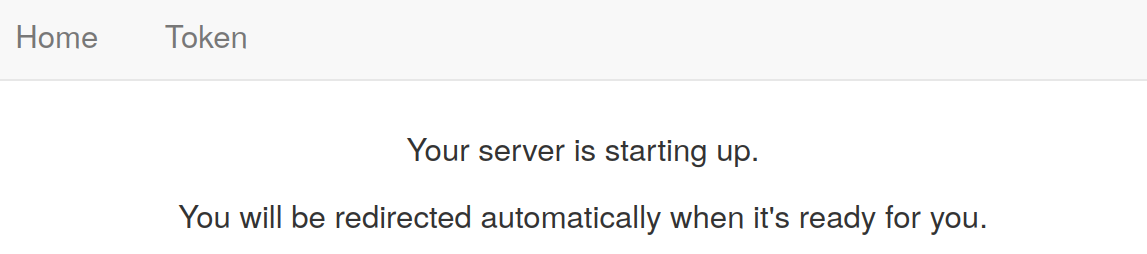
- After a successful start-up, you will see something like this. On the left are files - it is possible to upload (up arrow or drag and drop the files into this area) or create new ones. On the right are the windows for code, notebooks and other materials. In the example a new Jupyter Notebook has just been created.
- In a Jupyter Notebook it is possible to use Python or R. In the Notebook you need to pick the kernel you want to work with. This can be done when creating the Notebook or from the menu Kernel -> Change Kernel or clicking on the kernel name on the top right on an existing doucment. After this the following view will open.
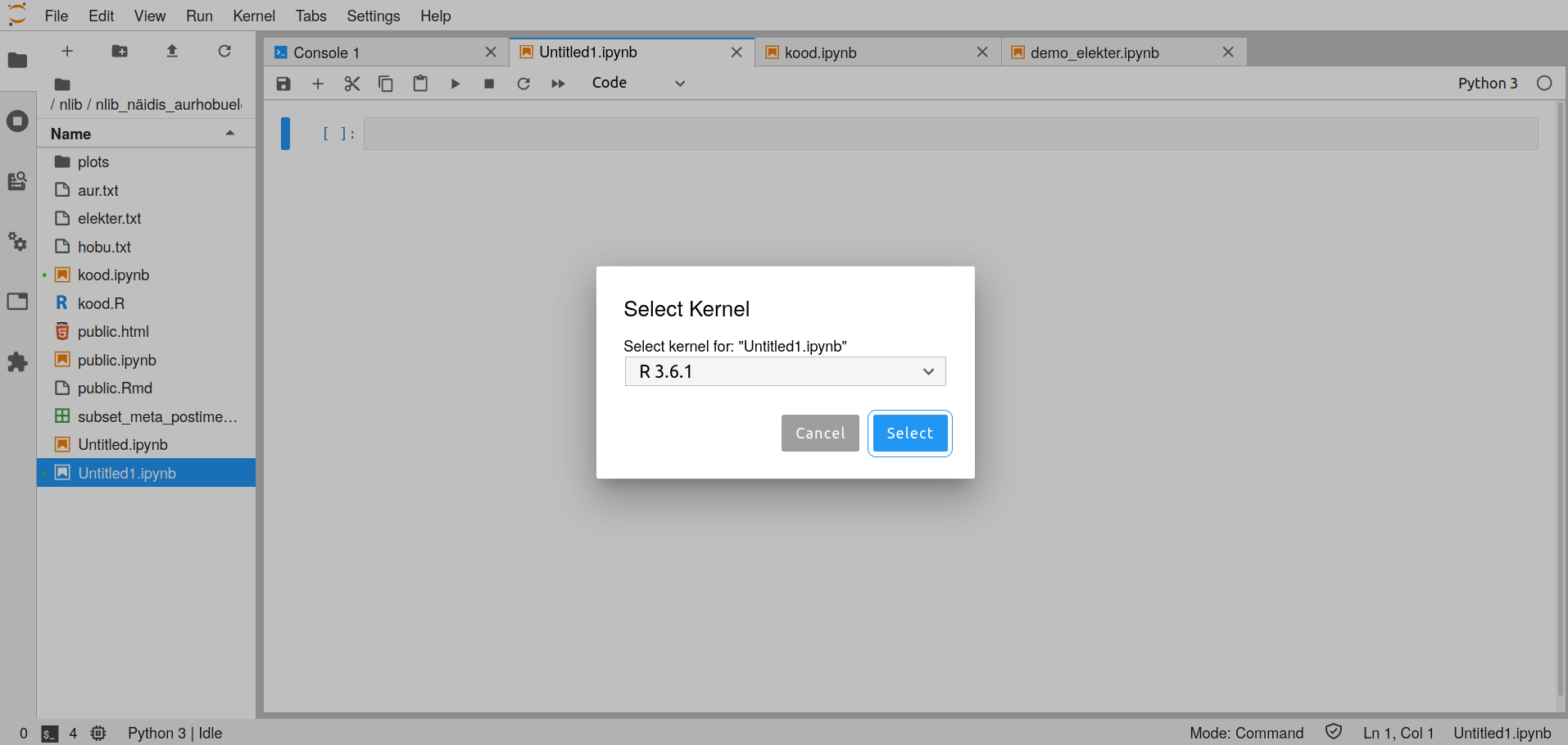
- The access to the texts at the library currently relies on R. It is recommended to do an initial query with this, and use your preferred tools after.
The package
The access to the files is supported by an R package digar.txts which uses a few simple commands to 1) get an overview of the data and the associated files, 2) form subsets of the data, 3) perform text searches on them, and 4) extract the immediate context of the text matches. The search results may for example be stored in a table and downladed as a smaller collection for offline use.
These commands are:
- get_digar_overvew() - gives overview of the collection (issue-level)
- get_subset_meta() - gives metainformation of the subset (article-level)
- do_subset_search() - does a search on the subset and prints results to file
- get_concordances() - gets word-in-context within the search results
Any R commands or packages can fit for further processing. While the JupyterHub environment supports both R and Python, but each Notebook is usually in only one of them.
Using code
- First, install the required package
#Install package remotes, if needed. JupyterLab should have it.
#install.packages("remotes")
#Since the JypiterLab that we use does not have write-access to
#all the files, we specify a local folder for our packages.
dir.create("R_pckg")
remotes::install_github("peeter-t2/digar.txts",lib="~/R_pckg/",upgrade="never")- Activate the package that was installed, use
library(digar.txts,lib.loc="~/R_pckg/")- Use get_digar_overview() to get overview of the collections (issue-level).
all_issues <- get_digar_overview()- Build a custom subset through any tools in R. Here is a tidyverse style example.
library(tidyverse)
subset <- all_issues %>%
filter(DocumentType=="NEWSPAPER") %>%
filter(year>1880&year<1940) %>%
filter(keyid=="postimeesew")
- Get meta information on that subset with get_subset_meta(). If this information is reused, sometimes storing the data is useful wth the commented lines. For complete metadata, we also need the earlier collection overview all_issues.
subset_meta <- get_subset_meta(subset)
#potentially write to file, for easier access if returning to it
#readr::write_tsv(subset_meta,"subset_meta_postimeesew1.tsv")
#subset_meta <- readr::read_tsv("subset_meta_postimeesew1.tsv")- Do a search with do_subset_search(). This exports the search results into a file as a set of articles. do_subset_search() ignores case.
do_subset_search(searchterm="lurich", searchfile="lurich1.txt",subset)- Read the search results. Use any R tools. It’s useful to name the id and text columns id and txt.
texts <- fread("lurich1.txt",header=F)[,.(id=V1,txt=V2)]- Get concordances using the get_concordances() command
concs <- get_concordances(searchterm="[Ll]urich",texts=texts,before=30,after=30,txt="txt",id="id")Note: to use ctrl+shift+m keyboard shortcut for the %>% pipe in Jupyter, do this. Add this code in Settings -> Advanced Settings Editor… -> Keyboard Shortcuts, on the left in the User Preferences box.
{
"shortcuts": [
{
"command": "notebook:replace-selection",
"selector": ".jp-Notebook",
"keys": ["Ctrl Shift M"],
"args": {"text": '%>% '}
}
]
}Simple R commands
- <- - store the values
- %>% - ‘pipe’ the values forward
- filter() - filter the data (preserving suitable values)
- count() - count the occurrences of the values
- mutate() - create a new column (e.g. n(), row_number(), min(), max() can be used to calculate new values
- head(n) - take the n first rows